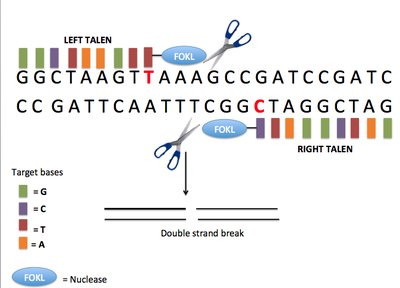
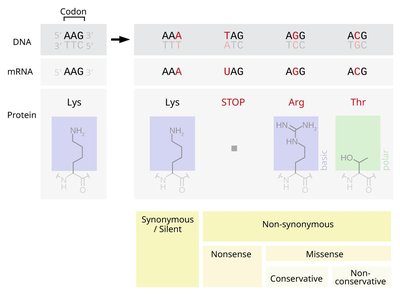
WITHDRAWN: Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model Animals Article Swipe
J. Wang
,
Jiaqi Wang
,
Alexandra Florea
,
Robert R. Stewart
,
Maeve Ballantyne
,
Carl S. Tucker
,
Ross D. Houston
,
Diego Robledo
·
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.04.25.650635
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.04.25.650635
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.04.25.650635
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1101/2025.04.25.650635
Statement for withdrawal The authors have withdrawn their manuscript because it was submitted and made public without full consent of all authors. Therefore, the authors do not wish this work to be cited as reference for the project. If you have any questions, please contact the corresponding author.
Related Topics
Concepts
Biology
Gene
Stop codon
Genetics
Genome
Computational biology
Genome editing
Start codon
Codon usage bias
Point mutation
Base pair
ENCODE
Function (biology)
Mutation
Model organism
Messenger RNA
Metadata
- Type
- preprint
- Language
- en
- Landing Page
- https://doi.org/10.1101/2025.04.25.650635
- https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdf
- OA Status
- green
- References
- 56
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4409953553
All OpenAlex metadata
Raw OpenAlex JSON
- OpenAlex ID
-
https://openalex.org/W4409953553Canonical identifier for this work in OpenAlex
- DOI
-
https://doi.org/10.1101/2025.04.25.650635Digital Object Identifier
- Title
-
WITHDRAWN: Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model AnimalsWork title
- Type
-
preprintOpenAlex work type
- Language
-
enPrimary language
- Publication year
-
2025Year of publication
- Publication date
-
2025-04-29Full publication date if available
- Authors
-
J. Wang, Jiaqi Wang, Alexandra Florea, Robert R. Stewart, Maeve Ballantyne, Carl S. Tucker, Ross D. Houston, Diego RobledoList of authors in order
- Landing page
-
https://doi.org/10.1101/2025.04.25.650635Publisher landing page
- PDF URL
-
https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdfDirect link to full text PDF
- Open access
-
YesWhether a free full text is available
- OA status
-
greenOpen access status per OpenAlex
- OA URL
-
https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdfDirect OA link when available
- Concepts
-
Biology, Gene, Stop codon, Genetics, Genome, Computational biology, Genome editing, Start codon, Codon usage bias, Point mutation, Base pair, ENCODE, Function (biology), Mutation, Model organism, Messenger RNATop concepts (fields/topics) attached by OpenAlex
- Cited by
-
0Total citation count in OpenAlex
- References (count)
-
56Number of works referenced by this work
- Related works (count)
-
10Other works algorithmically related by OpenAlex
Full payload
| id | https://openalex.org/W4409953553 |
|---|---|
| doi | https://doi.org/10.1101/2025.04.25.650635 |
| ids.doi | https://doi.org/10.1101/2025.04.25.650635 |
| ids.openalex | https://openalex.org/W4409953553 |
| fwci | 0.0 |
| type | preprint |
| title | WITHDRAWN: Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model Animals |
| biblio.issue | |
| biblio.volume | |
| biblio.last_page | |
| biblio.first_page | |
| topics[0].id | https://openalex.org/T10878 |
| topics[0].field.id | https://openalex.org/fields/13 |
| topics[0].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[0].score | 1.0 |
| topics[0].domain.id | https://openalex.org/domains/1 |
| topics[0].domain.display_name | Life Sciences |
| topics[0].subfield.id | https://openalex.org/subfields/1312 |
| topics[0].subfield.display_name | Molecular Biology |
| topics[0].display_name | CRISPR and Genetic Engineering |
| topics[1].id | https://openalex.org/T10521 |
| topics[1].field.id | https://openalex.org/fields/13 |
| topics[1].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[1].score | 0.9987999796867371 |
| topics[1].domain.id | https://openalex.org/domains/1 |
| topics[1].domain.display_name | Life Sciences |
| topics[1].subfield.id | https://openalex.org/subfields/1312 |
| topics[1].subfield.display_name | Molecular Biology |
| topics[1].display_name | RNA and protein synthesis mechanisms |
| topics[2].id | https://openalex.org/T10207 |
| topics[2].field.id | https://openalex.org/fields/13 |
| topics[2].field.display_name | Biochemistry, Genetics and Molecular Biology |
| topics[2].score | 0.994700014591217 |
| topics[2].domain.id | https://openalex.org/domains/1 |
| topics[2].domain.display_name | Life Sciences |
| topics[2].subfield.id | https://openalex.org/subfields/1312 |
| topics[2].subfield.display_name | Molecular Biology |
| topics[2].display_name | Advanced biosensing and bioanalysis techniques |
| is_xpac | False |
| apc_list | |
| apc_paid | |
| concepts[0].id | https://openalex.org/C86803240 |
| concepts[0].level | 0 |
| concepts[0].score | 0.6570309400558472 |
| concepts[0].wikidata | https://www.wikidata.org/wiki/Q420 |
| concepts[0].display_name | Biology |
| concepts[1].id | https://openalex.org/C104317684 |
| concepts[1].level | 2 |
| concepts[1].score | 0.6475759744644165 |
| concepts[1].wikidata | https://www.wikidata.org/wiki/Q7187 |
| concepts[1].display_name | Gene |
| concepts[2].id | https://openalex.org/C20580545 |
| concepts[2].level | 3 |
| concepts[2].score | 0.5921586155891418 |
| concepts[2].wikidata | https://www.wikidata.org/wiki/Q28354 |
| concepts[2].display_name | Stop codon |
| concepts[3].id | https://openalex.org/C54355233 |
| concepts[3].level | 1 |
| concepts[3].score | 0.5379930138587952 |
| concepts[3].wikidata | https://www.wikidata.org/wiki/Q7162 |
| concepts[3].display_name | Genetics |
| concepts[4].id | https://openalex.org/C141231307 |
| concepts[4].level | 3 |
| concepts[4].score | 0.5239384770393372 |
| concepts[4].wikidata | https://www.wikidata.org/wiki/Q7020 |
| concepts[4].display_name | Genome |
| concepts[5].id | https://openalex.org/C70721500 |
| concepts[5].level | 1 |
| concepts[5].score | 0.5199795365333557 |
| concepts[5].wikidata | https://www.wikidata.org/wiki/Q177005 |
| concepts[5].display_name | Computational biology |
| concepts[6].id | https://openalex.org/C144501496 |
| concepts[6].level | 4 |
| concepts[6].score | 0.5027058124542236 |
| concepts[6].wikidata | https://www.wikidata.org/wiki/Q5533489 |
| concepts[6].display_name | Genome editing |
| concepts[7].id | https://openalex.org/C176990463 |
| concepts[7].level | 4 |
| concepts[7].score | 0.5010049343109131 |
| concepts[7].wikidata | https://www.wikidata.org/wiki/Q1854186 |
| concepts[7].display_name | Start codon |
| concepts[8].id | https://openalex.org/C87253356 |
| concepts[8].level | 4 |
| concepts[8].score | 0.5009620189666748 |
| concepts[8].wikidata | https://www.wikidata.org/wiki/Q1106472 |
| concepts[8].display_name | Codon usage bias |
| concepts[9].id | https://openalex.org/C176944494 |
| concepts[9].level | 4 |
| concepts[9].score | 0.4962649941444397 |
| concepts[9].wikidata | https://www.wikidata.org/wiki/Q1415380 |
| concepts[9].display_name | Point mutation |
| concepts[10].id | https://openalex.org/C185581394 |
| concepts[10].level | 3 |
| concepts[10].score | 0.4533502459526062 |
| concepts[10].wikidata | https://www.wikidata.org/wiki/Q145911 |
| concepts[10].display_name | Base pair |
| concepts[11].id | https://openalex.org/C66746571 |
| concepts[11].level | 3 |
| concepts[11].score | 0.44161587953567505 |
| concepts[11].wikidata | https://www.wikidata.org/wiki/Q1134833 |
| concepts[11].display_name | ENCODE |
| concepts[12].id | https://openalex.org/C14036430 |
| concepts[12].level | 2 |
| concepts[12].score | 0.4258379340171814 |
| concepts[12].wikidata | https://www.wikidata.org/wiki/Q3736076 |
| concepts[12].display_name | Function (biology) |
| concepts[13].id | https://openalex.org/C501734568 |
| concepts[13].level | 3 |
| concepts[13].score | 0.42018482089042664 |
| concepts[13].wikidata | https://www.wikidata.org/wiki/Q42918 |
| concepts[13].display_name | Mutation |
| concepts[14].id | https://openalex.org/C19843653 |
| concepts[14].level | 3 |
| concepts[14].score | 0.4172777533531189 |
| concepts[14].wikidata | https://www.wikidata.org/wiki/Q213907 |
| concepts[14].display_name | Model organism |
| concepts[15].id | https://openalex.org/C105580179 |
| concepts[15].level | 3 |
| concepts[15].score | 0.31638145446777344 |
| concepts[15].wikidata | https://www.wikidata.org/wiki/Q188928 |
| concepts[15].display_name | Messenger RNA |
| keywords[0].id | https://openalex.org/keywords/biology |
| keywords[0].score | 0.6570309400558472 |
| keywords[0].display_name | Biology |
| keywords[1].id | https://openalex.org/keywords/gene |
| keywords[1].score | 0.6475759744644165 |
| keywords[1].display_name | Gene |
| keywords[2].id | https://openalex.org/keywords/stop-codon |
| keywords[2].score | 0.5921586155891418 |
| keywords[2].display_name | Stop codon |
| keywords[3].id | https://openalex.org/keywords/genetics |
| keywords[3].score | 0.5379930138587952 |
| keywords[3].display_name | Genetics |
| keywords[4].id | https://openalex.org/keywords/genome |
| keywords[4].score | 0.5239384770393372 |
| keywords[4].display_name | Genome |
| keywords[5].id | https://openalex.org/keywords/computational-biology |
| keywords[5].score | 0.5199795365333557 |
| keywords[5].display_name | Computational biology |
| keywords[6].id | https://openalex.org/keywords/genome-editing |
| keywords[6].score | 0.5027058124542236 |
| keywords[6].display_name | Genome editing |
| keywords[7].id | https://openalex.org/keywords/start-codon |
| keywords[7].score | 0.5010049343109131 |
| keywords[7].display_name | Start codon |
| keywords[8].id | https://openalex.org/keywords/codon-usage-bias |
| keywords[8].score | 0.5009620189666748 |
| keywords[8].display_name | Codon usage bias |
| keywords[9].id | https://openalex.org/keywords/point-mutation |
| keywords[9].score | 0.4962649941444397 |
| keywords[9].display_name | Point mutation |
| keywords[10].id | https://openalex.org/keywords/base-pair |
| keywords[10].score | 0.4533502459526062 |
| keywords[10].display_name | Base pair |
| keywords[11].id | https://openalex.org/keywords/encode |
| keywords[11].score | 0.44161587953567505 |
| keywords[11].display_name | ENCODE |
| keywords[12].id | https://openalex.org/keywords/function |
| keywords[12].score | 0.4258379340171814 |
| keywords[12].display_name | Function (biology) |
| keywords[13].id | https://openalex.org/keywords/mutation |
| keywords[13].score | 0.42018482089042664 |
| keywords[13].display_name | Mutation |
| keywords[14].id | https://openalex.org/keywords/model-organism |
| keywords[14].score | 0.4172777533531189 |
| keywords[14].display_name | Model organism |
| keywords[15].id | https://openalex.org/keywords/messenger-rna |
| keywords[15].score | 0.31638145446777344 |
| keywords[15].display_name | Messenger RNA |
| language | en |
| locations[0].id | doi:10.1101/2025.04.25.650635 |
| locations[0].is_oa | True |
| locations[0].source.id | https://openalex.org/S4306402567 |
| locations[0].source.issn | |
| locations[0].source.type | repository |
| locations[0].source.is_oa | False |
| locations[0].source.issn_l | |
| locations[0].source.is_core | False |
| locations[0].source.is_in_doaj | False |
| locations[0].source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| locations[0].source.host_organization | https://openalex.org/I2750212522 |
| locations[0].source.host_organization_name | Cold Spring Harbor Laboratory |
| locations[0].source.host_organization_lineage | https://openalex.org/I2750212522 |
| locations[0].license | |
| locations[0].pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdf |
| locations[0].version | acceptedVersion |
| locations[0].raw_type | posted-content |
| locations[0].license_id | |
| locations[0].is_accepted | True |
| locations[0].is_published | False |
| locations[0].raw_source_name | |
| locations[0].landing_page_url | https://doi.org/10.1101/2025.04.25.650635 |
| indexed_in | crossref |
| authorships[0].author.id | https://openalex.org/A5023966190 |
| authorships[0].author.orcid | https://orcid.org/0009-0000-5240-4574 |
| authorships[0].author.display_name | J. Wang |
| authorships[0].author_position | first |
| authorships[0].raw_author_name | Jinhai Wang |
| authorships[0].is_corresponding | False |
| authorships[1].author.id | https://openalex.org/A5064769317 |
| authorships[1].author.orcid | https://orcid.org/0000-0003-3423-1328 |
| authorships[1].author.display_name | Jiaqi Wang |
| authorships[1].author_position | middle |
| authorships[1].raw_author_name | Jiaqi Wang |
| authorships[1].is_corresponding | False |
| authorships[2].author.id | https://openalex.org/A5114294819 |
| authorships[2].author.orcid | |
| authorships[2].author.display_name | Alexandra Florea |
| authorships[2].author_position | middle |
| authorships[2].raw_author_name | Alexandra Florea |
| authorships[2].is_corresponding | False |
| authorships[3].author.id | https://openalex.org/A5020914067 |
| authorships[3].author.orcid | https://orcid.org/0000-0003-1694-9395 |
| authorships[3].author.display_name | Robert R. Stewart |
| authorships[3].author_position | middle |
| authorships[3].raw_author_name | Robert Stewart |
| authorships[3].is_corresponding | False |
| authorships[4].author.id | https://openalex.org/A5028627100 |
| authorships[4].author.orcid | https://orcid.org/0000-0002-3388-6261 |
| authorships[4].author.display_name | Maeve Ballantyne |
| authorships[4].author_position | middle |
| authorships[4].raw_author_name | Maeve Ballantyne |
| authorships[4].is_corresponding | False |
| authorships[5].author.id | https://openalex.org/A5105938841 |
| authorships[5].author.orcid | |
| authorships[5].author.display_name | Carl S. Tucker |
| authorships[5].author_position | middle |
| authorships[5].raw_author_name | Carl Tucker |
| authorships[5].is_corresponding | False |
| authorships[6].author.id | https://openalex.org/A5005051455 |
| authorships[6].author.orcid | https://orcid.org/0000-0003-1805-0762 |
| authorships[6].author.display_name | Ross D. Houston |
| authorships[6].author_position | middle |
| authorships[6].raw_author_name | Ross D. Houston |
| authorships[6].is_corresponding | False |
| authorships[7].author.id | https://openalex.org/A5071973746 |
| authorships[7].author.orcid | https://orcid.org/0000-0002-9616-5912 |
| authorships[7].author.display_name | Diego Robledo |
| authorships[7].author_position | last |
| authorships[7].raw_author_name | Diego Robledo |
| authorships[7].is_corresponding | False |
| has_content.pdf | True |
| has_content.grobid_xml | False |
| is_paratext | False |
| open_access.is_oa | True |
| open_access.oa_url | https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdf |
| open_access.oa_status | green |
| open_access.any_repository_has_fulltext | False |
| created_date | 2025-04-30T00:00:00 |
| display_name | WITHDRAWN: Codon-Optimized Base Editors Enable Efficient Base Substitution in Non-model Animals |
| has_fulltext | False |
| is_retracted | False |
| updated_date | 2025-11-06T03:46:38.306776 |
| primary_topic.id | https://openalex.org/T10878 |
| primary_topic.field.id | https://openalex.org/fields/13 |
| primary_topic.field.display_name | Biochemistry, Genetics and Molecular Biology |
| primary_topic.score | 1.0 |
| primary_topic.domain.id | https://openalex.org/domains/1 |
| primary_topic.domain.display_name | Life Sciences |
| primary_topic.subfield.id | https://openalex.org/subfields/1312 |
| primary_topic.subfield.display_name | Molecular Biology |
| primary_topic.display_name | CRISPR and Genetic Engineering |
| related_works | https://openalex.org/W2054043430, https://openalex.org/W2137401535, https://openalex.org/W2028165934, https://openalex.org/W1793475955, https://openalex.org/W2162549018, https://openalex.org/W2100776225, https://openalex.org/W2082497884, https://openalex.org/W2167973314, https://openalex.org/W2912466504, https://openalex.org/W1998747977 |
| cited_by_count | 0 |
| locations_count | 1 |
| best_oa_location.id | doi:10.1101/2025.04.25.650635 |
| best_oa_location.is_oa | True |
| best_oa_location.source.id | https://openalex.org/S4306402567 |
| best_oa_location.source.issn | |
| best_oa_location.source.type | repository |
| best_oa_location.source.is_oa | False |
| best_oa_location.source.issn_l | |
| best_oa_location.source.is_core | False |
| best_oa_location.source.is_in_doaj | False |
| best_oa_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| best_oa_location.source.host_organization | https://openalex.org/I2750212522 |
| best_oa_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| best_oa_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| best_oa_location.license | |
| best_oa_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdf |
| best_oa_location.version | acceptedVersion |
| best_oa_location.raw_type | posted-content |
| best_oa_location.license_id | |
| best_oa_location.is_accepted | True |
| best_oa_location.is_published | False |
| best_oa_location.raw_source_name | |
| best_oa_location.landing_page_url | https://doi.org/10.1101/2025.04.25.650635 |
| primary_location.id | doi:10.1101/2025.04.25.650635 |
| primary_location.is_oa | True |
| primary_location.source.id | https://openalex.org/S4306402567 |
| primary_location.source.issn | |
| primary_location.source.type | repository |
| primary_location.source.is_oa | False |
| primary_location.source.issn_l | |
| primary_location.source.is_core | False |
| primary_location.source.is_in_doaj | False |
| primary_location.source.display_name | bioRxiv (Cold Spring Harbor Laboratory) |
| primary_location.source.host_organization | https://openalex.org/I2750212522 |
| primary_location.source.host_organization_name | Cold Spring Harbor Laboratory |
| primary_location.source.host_organization_lineage | https://openalex.org/I2750212522 |
| primary_location.license | |
| primary_location.pdf_url | https://www.biorxiv.org/content/biorxiv/early/2025/04/29/2025.04.25.650635.full.pdf |
| primary_location.version | acceptedVersion |
| primary_location.raw_type | posted-content |
| primary_location.license_id | |
| primary_location.is_accepted | True |
| primary_location.is_published | False |
| primary_location.raw_source_name | |
| primary_location.landing_page_url | https://doi.org/10.1101/2025.04.25.650635 |
| publication_date | 2025-04-29 |
| publication_year | 2025 |
| referenced_works | https://openalex.org/W3035764705, https://openalex.org/W2115603949, https://openalex.org/W3165675334, https://openalex.org/W2157584580, https://openalex.org/W4225695788, https://openalex.org/W4399785830, https://openalex.org/W1971191097, https://openalex.org/W4392159460, https://openalex.org/W2050480410, https://openalex.org/W2766599608, https://openalex.org/W2964213295, https://openalex.org/W3087609775, https://openalex.org/W3032652943, https://openalex.org/W4295013996, https://openalex.org/W4300426741, https://openalex.org/W4404824804, https://openalex.org/W3017265939, https://openalex.org/W3121977414, https://openalex.org/W2883250809, https://openalex.org/W1555684022, https://openalex.org/W3194942572, https://openalex.org/W2045435533, https://openalex.org/W2809042973, https://openalex.org/W2336828812, https://openalex.org/W3119813698, https://openalex.org/W1969869269, https://openalex.org/W2030994161, https://openalex.org/W3153653697, https://openalex.org/W2107277218, https://openalex.org/W4283836463, https://openalex.org/W3165885441, https://openalex.org/W3162223676, https://openalex.org/W3093469725, https://openalex.org/W3088904989, https://openalex.org/W4389833780, https://openalex.org/W2256756668, https://openalex.org/W2897295193, https://openalex.org/W4282937657, https://openalex.org/W4385286090, https://openalex.org/W2039705224, https://openalex.org/W2545963238, https://openalex.org/W1974343902, https://openalex.org/W3013504322, https://openalex.org/W4391430084, https://openalex.org/W4404445114, https://openalex.org/W2411435798, https://openalex.org/W2892987358, https://openalex.org/W2954739215, https://openalex.org/W4223601921, https://openalex.org/W4323048053, https://openalex.org/W4404041312, https://openalex.org/W3109270346, https://openalex.org/W3039582431, https://openalex.org/W2921695994, https://openalex.org/W2893791662, https://openalex.org/W2916364858 |
| referenced_works_count | 56 |
| abstract_inverted_index.If | 38 |
| abstract_inverted_index.as | 33 |
| abstract_inverted_index.be | 31 |
| abstract_inverted_index.do | 25 |
| abstract_inverted_index.it | 10 |
| abstract_inverted_index.of | 19 |
| abstract_inverted_index.to | 30 |
| abstract_inverted_index.The | 3 |
| abstract_inverted_index.all | 20 |
| abstract_inverted_index.and | 13 |
| abstract_inverted_index.any | 41 |
| abstract_inverted_index.for | 1, 35 |
| abstract_inverted_index.not | 26 |
| abstract_inverted_index.the | 23, 36, 45 |
| abstract_inverted_index.was | 11 |
| abstract_inverted_index.you | 39 |
| abstract_inverted_index.full | 17 |
| abstract_inverted_index.have | 5, 40 |
| abstract_inverted_index.made | 14 |
| abstract_inverted_index.this | 28 |
| abstract_inverted_index.wish | 27 |
| abstract_inverted_index.work | 29 |
| abstract_inverted_index.cited | 32 |
| abstract_inverted_index.their | 7 |
| abstract_inverted_index.please | 43 |
| abstract_inverted_index.public | 15 |
| abstract_inverted_index.author. | 47 |
| abstract_inverted_index.authors | 4, 24 |
| abstract_inverted_index.because | 9 |
| abstract_inverted_index.consent | 18 |
| abstract_inverted_index.contact | 44 |
| abstract_inverted_index.without | 16 |
| abstract_inverted_index.authors. | 21 |
| abstract_inverted_index.project. | 37 |
| abstract_inverted_index.Statement | 0 |
| abstract_inverted_index.reference | 34 |
| abstract_inverted_index.submitted | 12 |
| abstract_inverted_index.withdrawn | 6 |
| abstract_inverted_index.Therefore, | 22 |
| abstract_inverted_index.manuscript | 8 |
| abstract_inverted_index.questions, | 42 |
| abstract_inverted_index.withdrawal | 2 |
| abstract_inverted_index.corresponding | 46 |
| cited_by_percentile_year | |
| countries_distinct_count | 0 |
| institutions_distinct_count | 8 |
| sustainable_development_goals[0].id | https://metadata.un.org/sdg/14 |
| sustainable_development_goals[0].score | 0.49000000953674316 |
| sustainable_development_goals[0].display_name | Life below water |
| citation_normalized_percentile.value | 0.14120542 |
| citation_normalized_percentile.is_in_top_1_percent | False |
| citation_normalized_percentile.is_in_top_10_percent | False |