Computational analysis to comprehend the structure-function properties of fibrinolytic enzymes from Bacillus spp for their efficient integration into industrial applications Article Swipe
 YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1016/j.heliyon.2024.e33895
YOU?
·
· 2024
· Open Access
·
· DOI: https://doi.org/10.1016/j.heliyon.2024.e33895
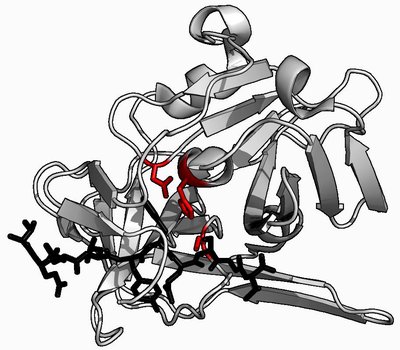
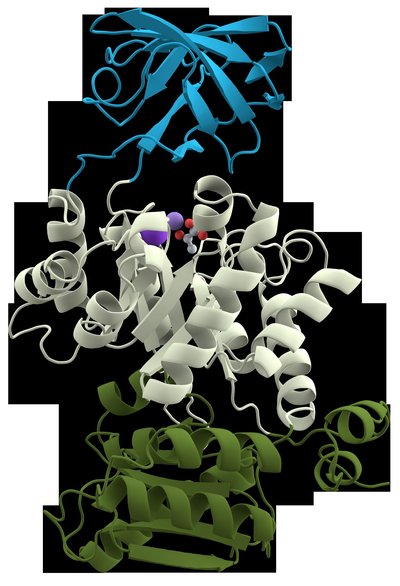
The alignment of sixty fibrinolytic serine protease enzymes (molecular mass 12-86 kDa) sequences showed 49 enzymes possess a conserved domain with a catalytic triad of Asp196, His242, and Ser569. The predicted instability and aliphatic indexes were 1.94-37.77, and 68.9-93.41, respectively, indicating high thermostability. The random coil means value suggested the predominance of this secondary structure in these proteases. A set of 50 amino acid residues representing motif 3 signifies the Peptidase S8/S53 domain that was invariably observed in 56 sequences. Additionally, 28 sequences have transmembrane helices, with two having the most disordered areas, and they pose 25 enzyme cleavage sites. A comparative analysis of the experimental work with the results of in-silico study put forward the characteristics of the enzyme sequences JF739176.1 and MF677779.1 to be considered when creating a potential mutant enzyme as these sequences are stable at high pH with thermostability and to exhibit αβ-fibrinogenase activity in both experimental and in-silico studies.
Related Topics To Compare & Contrast
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1016/j.heliyon.2024.e33895
- http://www.cell.com/article/S2405844024099262/pdf
- OA Status
- gold
- Cited By
- 6
- References
- 151
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4400191616