Predicting protein stability changes upon mutations with dual-view ensemble learning from single sequence Article Swipe
 YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbaf319
YOU?
·
· 2025
· Open Access
·
· DOI: https://doi.org/10.1093/bib/bbaf319
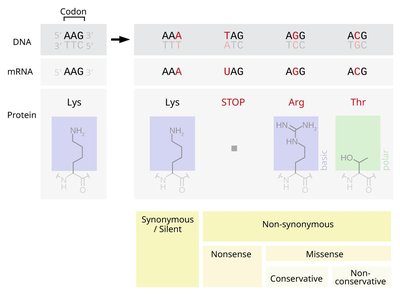
Predicting the protein stability changes upon mutations is one of the effective ways to improve the efficiency of protein engineering. Here, we propose a dual-view ensemble learning-based framework, DVE-stability, for mutation-induced protein stability change prediction from single sequence. DVE-stability integrates the global and local dependencies of mutations to capture the intramolecular interactions from two views through ensemble learning, in which a structural microenvironment simulation module is designed to indirectly introduce the information of structural microenvironment at the sequence level. DVE-stability achieved state-of-the-art prediction performance on seven single-point mutation benchmark datasets, and comprehensively surpassed other methods on five of them. Furthermore, DVE-stability outperformed other methods comprehensively through zero-shot inference on multiple-point mutation prediction task, demonstrating superior model generalizability to capture the epistasis of multiple-point mutations. More importantly, DVE-stability exhibited superior generalization performance in predicting rare beneficial mutations that are crucial for practical protein directed evolution scenarios. In addition, DVE-stability identified important intramolecular interactions via attention scores, demonstrating interpretable. Overall, DVE-stability provides a flexible and efficient tool for mutation-induced protein stability change prediction in an interpretable ensemble learning manner.
Related Topics To Compare & Contrast
- Type
- article
- Language
- en
- Landing Page
- https://doi.org/10.1093/bib/bbaf319
- https://academic.oup.com/bib/article-pdf/26/4/bbaf319/63725431/bbaf319.pdf
- OA Status
- gold
- References
- 65
- Related Works
- 10
- OpenAlex ID
- https://openalex.org/W4412184079